Molecule is an open source framework that is easy to use for validating and auditing code. It can be easily introduced into the CI-CD pipeline, thus keeping Ansible scripts relevant.
DevOps teams rely on ‘Infrastructure as a Code’ (IaaC) for productivity gains. Automation and speed are prerequisites in the cloud environment, where resources are identified by cattle nomenclature rather than pet nomenclature due to the sheer volumes.
Ansible is one of the leading technologies in the IaaC space. Its declarative style, ease of parameterisation and the availability of numerous modules make it the preferred framework to work with.
Any code, if not tested regularly, gets outdated and becomes irrelevant over time, and the same applies to Ansible. Daily testing is a best practice and must be introduced for Ansible scripts too—for example, keeping track of the latest version of a particular software during provisioning. Similarly, the dependency management repository used by apt and Yum may have broken dependencies due to changes. Scripts may also fail if a dependent URL is not available. Those working with Ansible would have faced these challenges.
This is where we introduce unit testing, which can run during the nightly build and can detect these failures well in advance. The Molecule project is a useful framework to introduce unit testing into Ansible code. One can use containers to test the individual role or use an array of containers to test complex deployments. Docker containers are useful, as they can save engineers from spawning multiple instances or using resource-hogging VMs in the cloud or on test machines. Docker is a lightweight technology which is used to verify the end state of the system, and after the test, the provisioned resources are destroyed thus cleaning up the environment.
Installing and working with Molecule is simple. Follow the steps shown below. First, get the OS updated as follows:
sudo apt-get update && sudo apt-get -y upgrade
Next, install Docker:
sudo apt install docker.io
Now install Molecule with the help of Python Pip:
sudo apt-get install python-pip python-dev build-essential sudo pip install --upgrade pip sudo pip install molecule
After the install, do a version check of Molecule. If the molecule version is not the latest, upgrade it as follows:
sudo pip install --upgrade molecule
It is always good to work with the latest version of Molecule as there are significant changes compared to an earlier version. Enabling or disabling modules in the latest version of Molecule is more effective. For example, a common problem faced is the forced audit errors that make Molecule fail. When starting to test with Molecule, at times audit errors can pose a roadblock. Disabling the Lint module during the initial phase can give you some speed to concentrate on writing tests rather than trying to fix the audit errors.
Here are a few features of Molecule explained, though the full toolset offers more.
1. Create: This creates a virtualised provider, which in our case is the Docker container.
2. Converge: Uses the provisioner and runs the Ansible scripts to the target running Docker containers.
3. Idempotency: Uses the provisioner to check the idempotency of Ansible scripts.
4. Lint: This does code audits of Ansible scripts, test code, test scripts, etc.
5. Verify: Runs the test scripts written.
6. Test: Runs the full steps needed for Molecule, i.e., create, converge, Lint, verify and destroy.
The roles need to be initialised by the following command:
molecule init role –role-name abc
This will create all the folders needed to create the role, and is similar to the following command:
ansible-galaxy init abc
There are certain differences in the directory structure, and some amount of manual folder creation may be required.
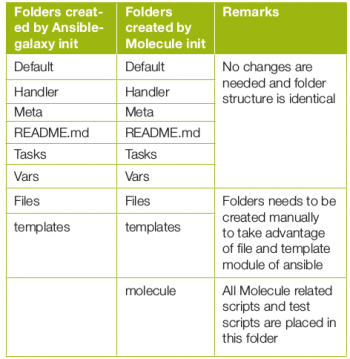
The Molecule folder has files in which one can put in a pre-created Molecule playbook and test scripts. The environment that is created is named default. This can be changed as per the project’s requirements.
One file that will be of interest is molecule.yml, which is placed at:
./molecule/default/molecule.yml
Another file which describes the playbook for the role is playbook.yml placed at:
./molecule/default/playbook.yml
Note: Molecule initialises the environment called default but engineers can use a name as per the environment used in the project.
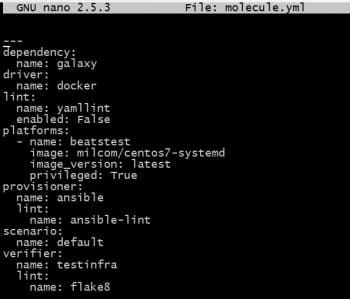
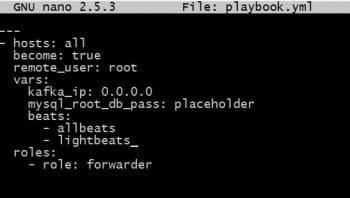
The sample Molecule file is shown in Figure 1. One needs to modify it as per the project’s requirement.
Note: Docker images used for testing need to be systemd enabled and should have privileges.
Take a look at the playbook. Make sure that you have made all the changes needed for all the parameters to be passed to the role.
Now we are ready to write test scripts using TestInfra (http://testinfra.readthedocs.io/en/latest/).
We need to go to the folder Molecule/default/tests. You can create a test assertion.
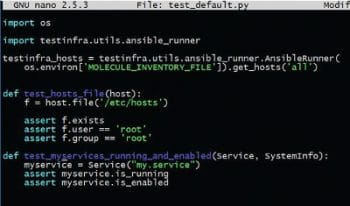
Test assertion is one of the most important steps in a testing framework. Molecule uses TestInfra (http://testinfra.readthedocs.io/en/latest) as its test validation framework. The tests are located in the Python file located at ./molecule/default/tests. The file contains assertions as shown in Figure 3.
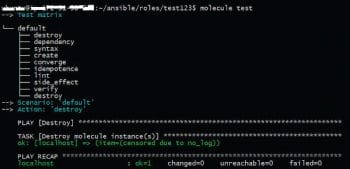
As can be seen, these assertions are declarative; hence it requires no effort to add them. The overall test is run with the Molecule test command as shown in Figure 4. It runs steps in an opinionated way—cleaning and provisioning the infrastructure, checking idempotency, testing assertions and at the end, cleaning resources when the tests are finished. Molecule can be extended to test complex scenarios and distributed clustered environments, too, in test machines.
As seen from the steps, Molecule can spin up a Docker container, do all the testing required and destroy the container after cleaning the infrastructure.